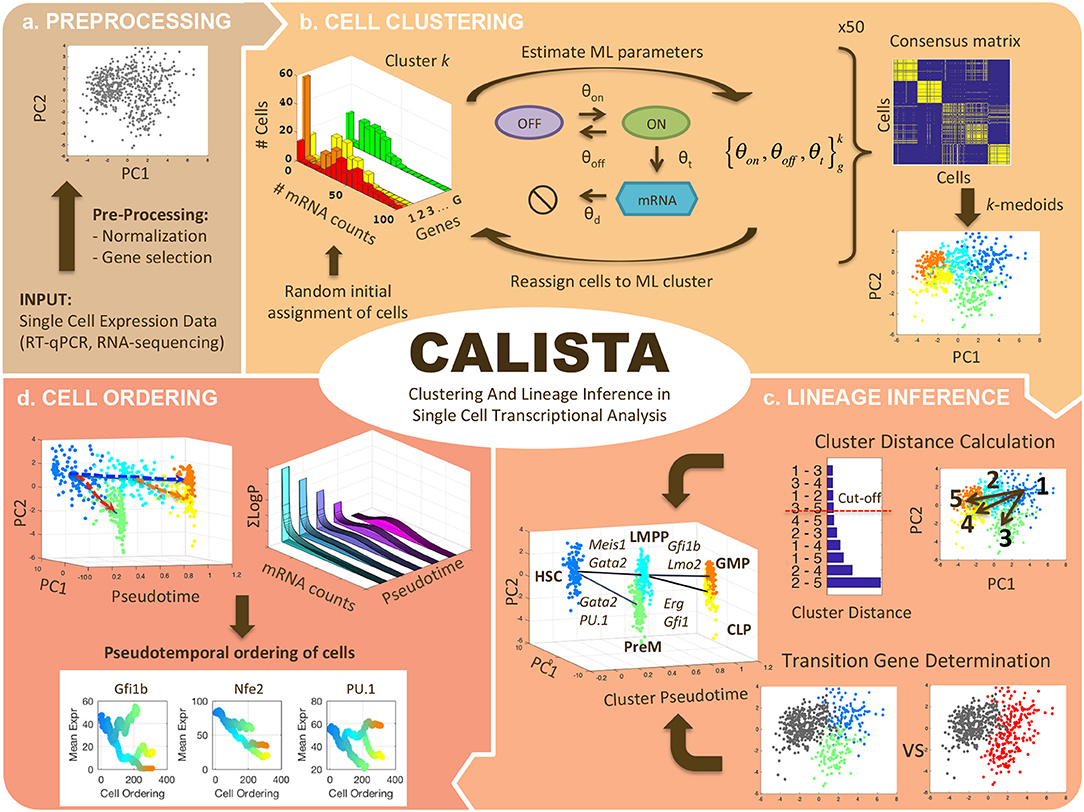
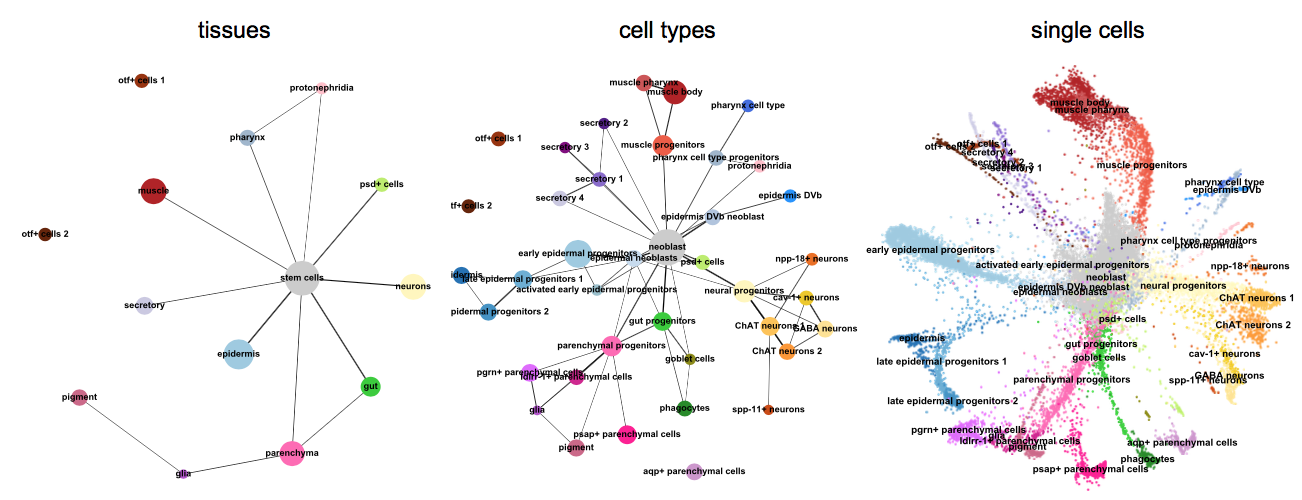
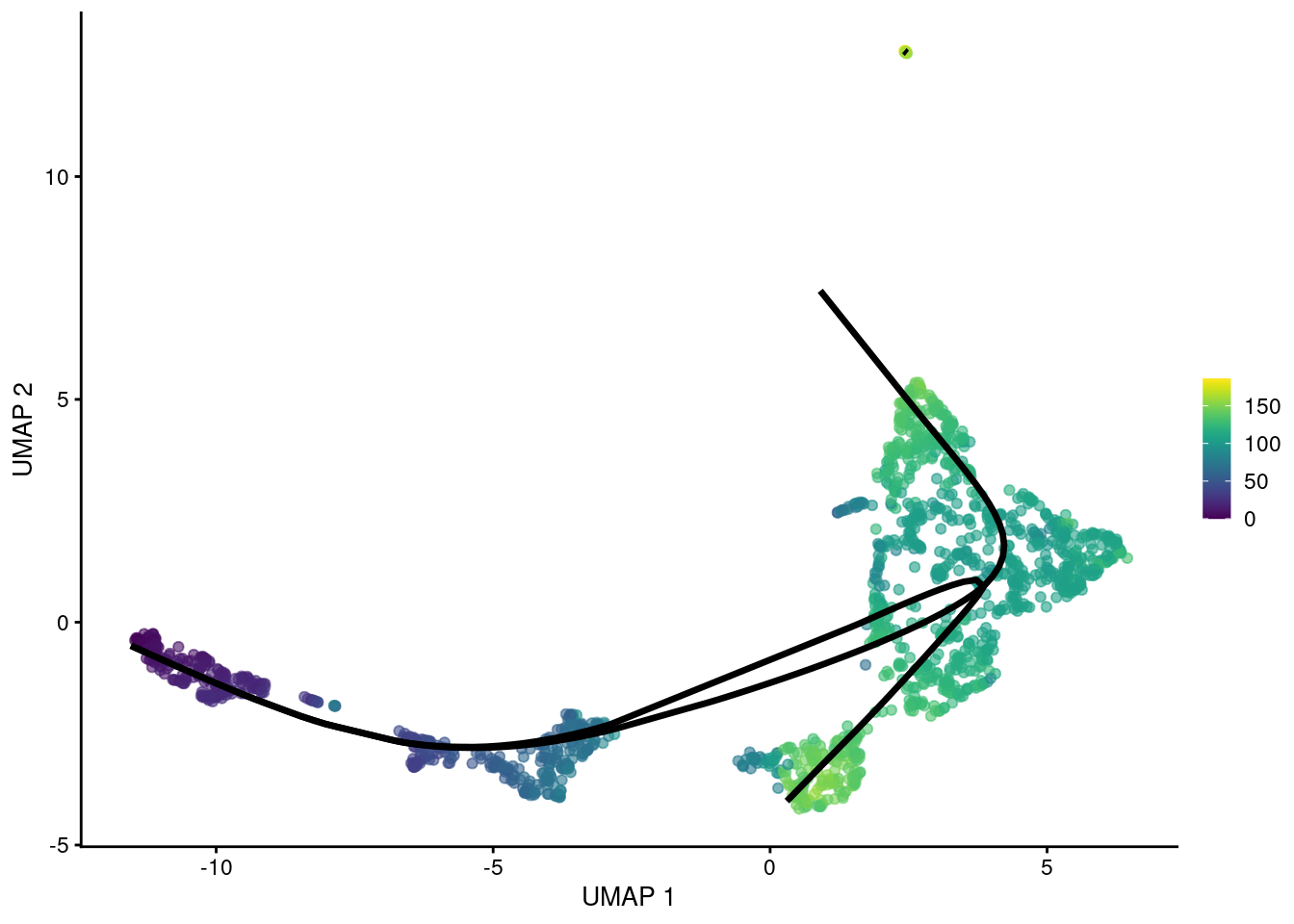
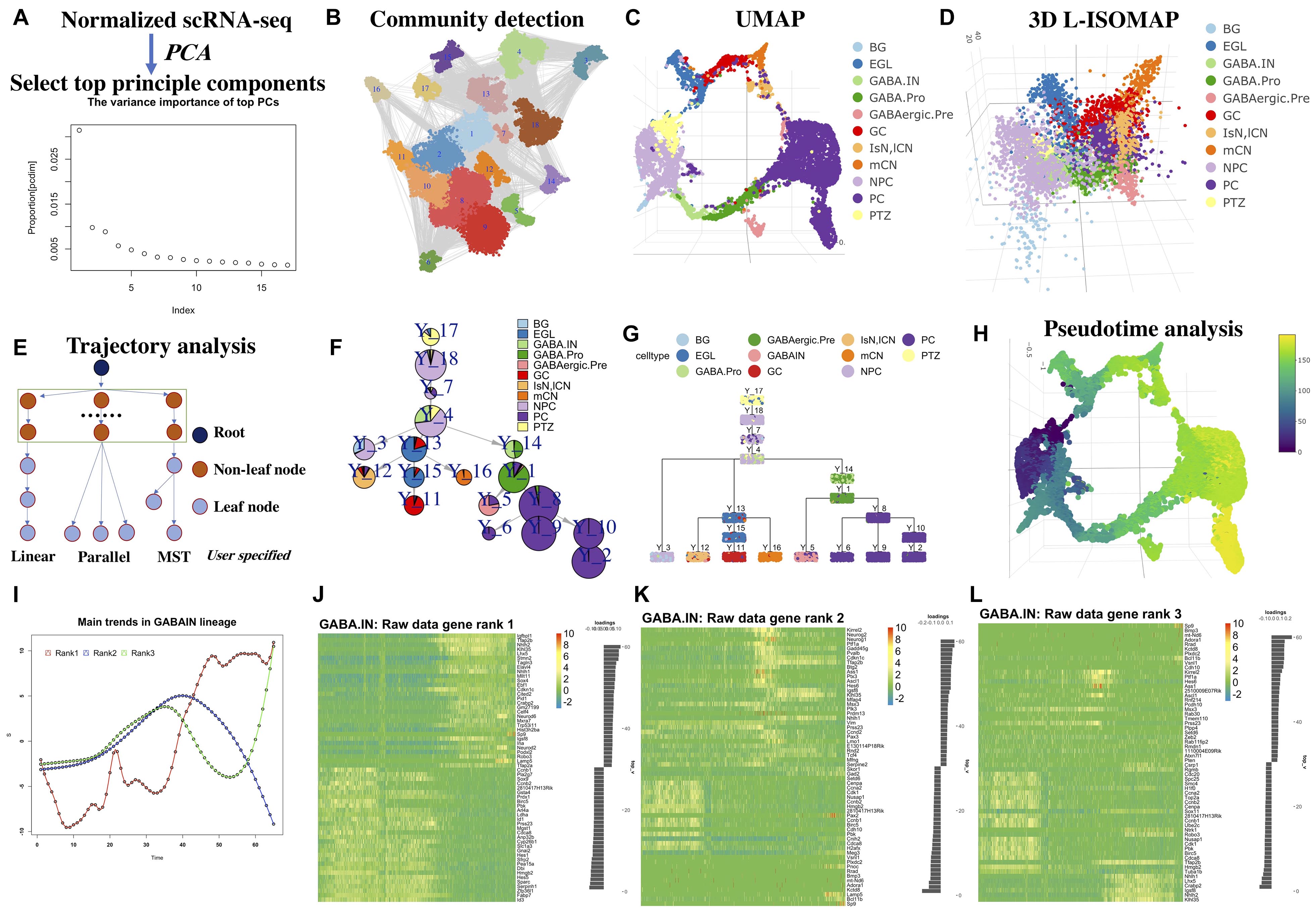
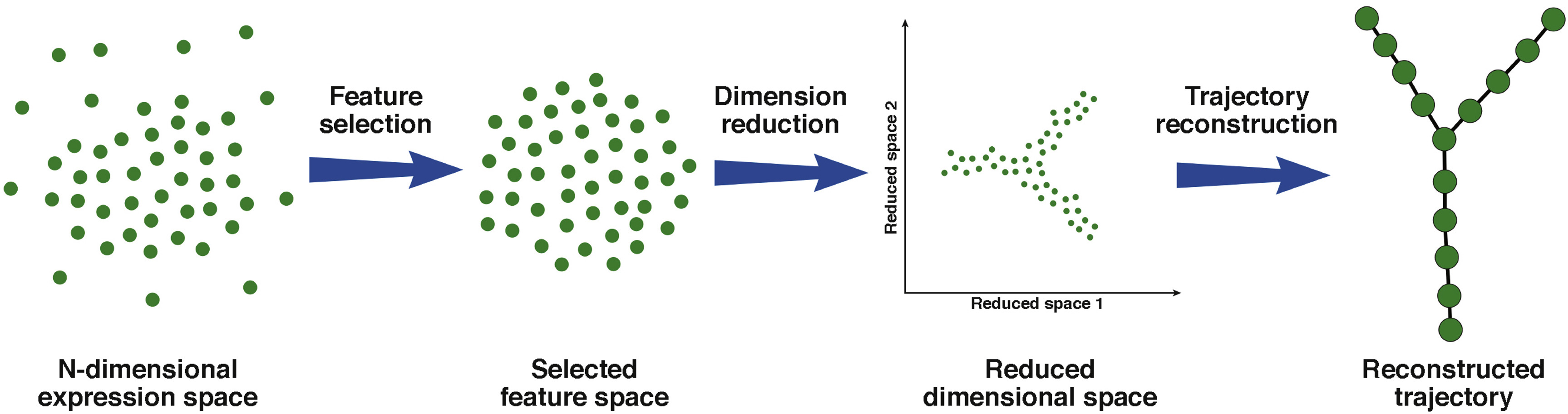
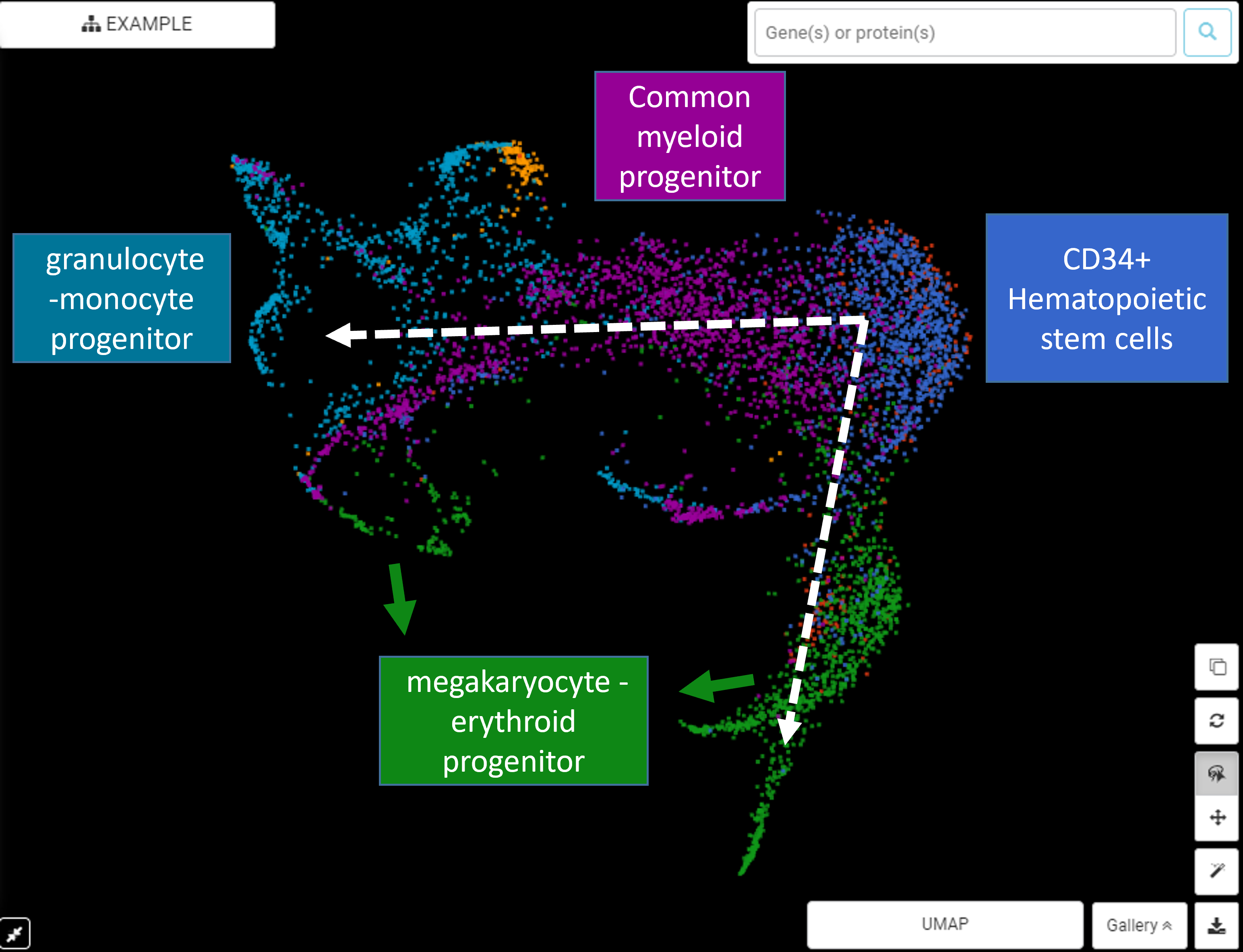
Unsupervised Trajectory Analysis of Single-Cell RNA-Seq and Imaging Data Reveals Alternative Tuft Cell Origins in the Gut: Cell Systems
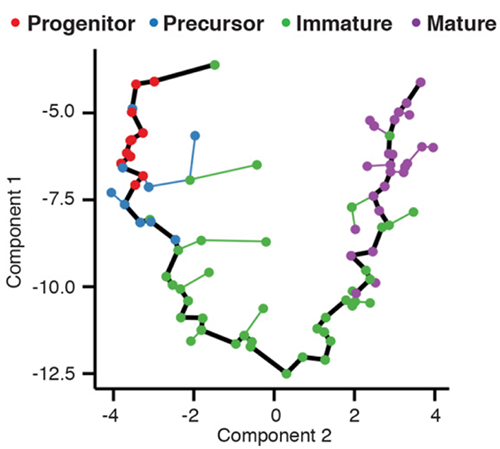
Single-Cell Trajectory Analysis Using Monocle 2 and scGPS Pseudotime... | Download Scientific Diagram

PseudotimeDE, statistical method that outperforms existing methods in its detection power | UCLA Computational Medicine
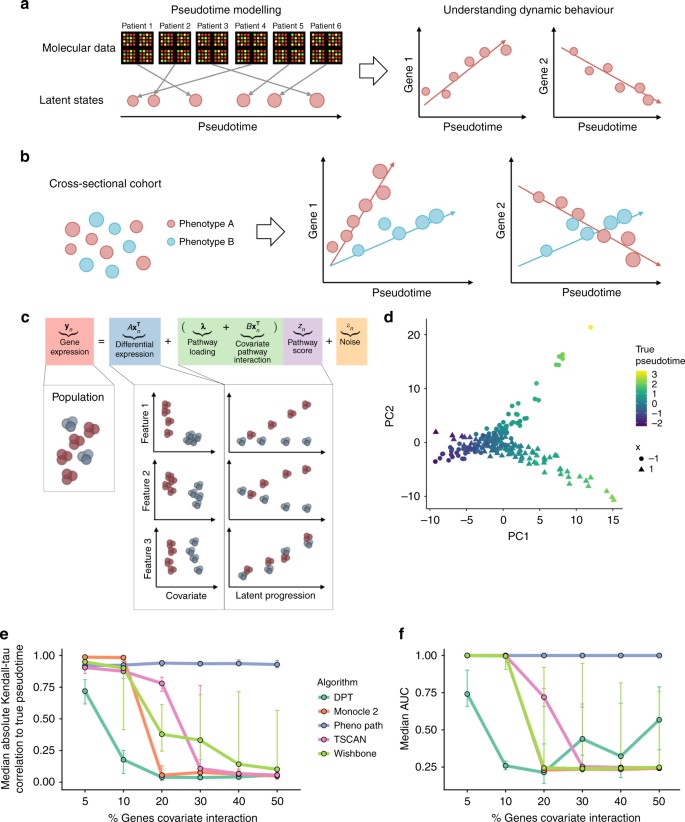
Uncovering pseudotemporal trajectories with covariates from single cell and bulk expression data | Nature Communications
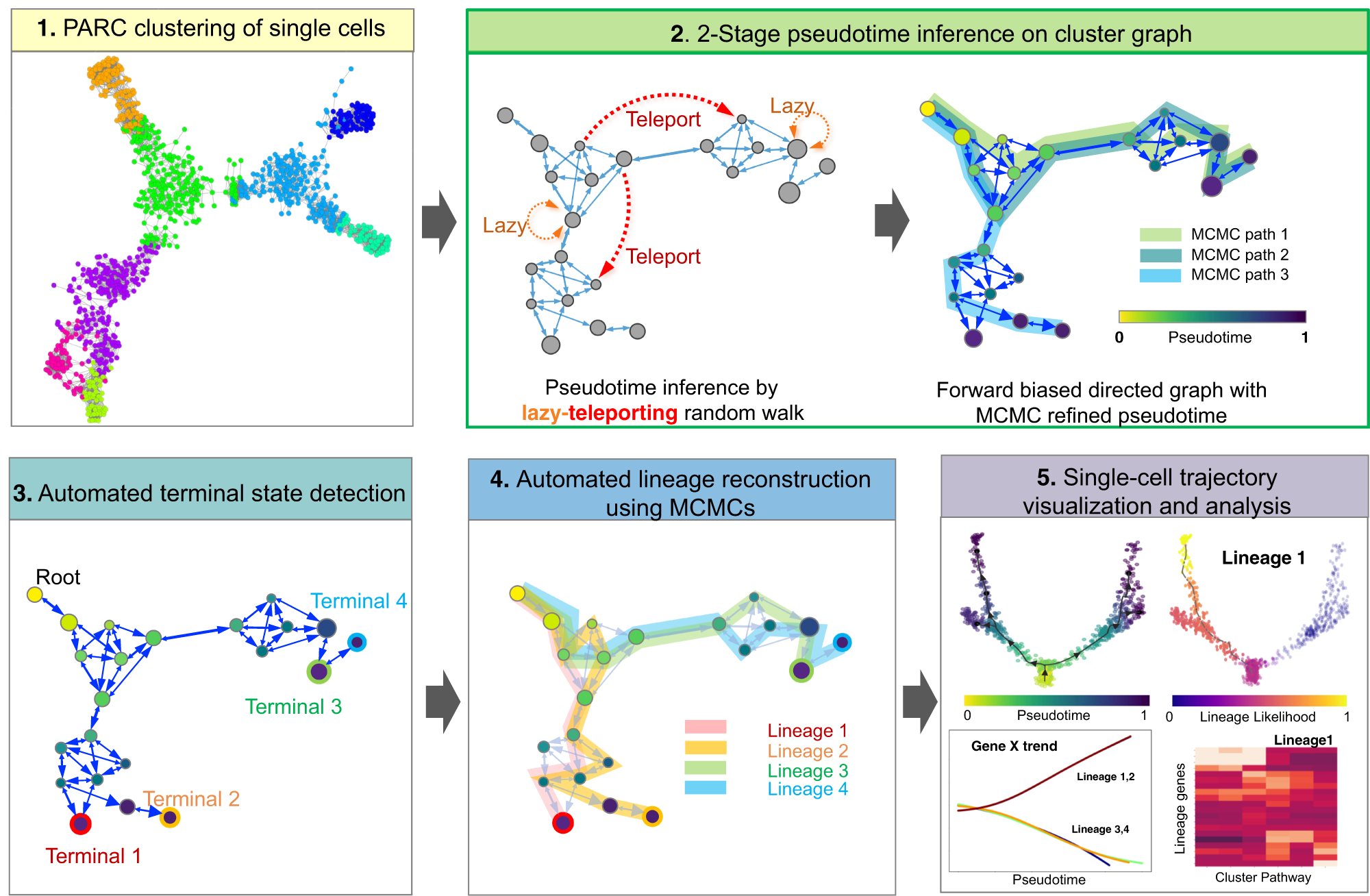
Generalized and scalable trajectory inference in single-cell omics data with VIA | Nature Communications
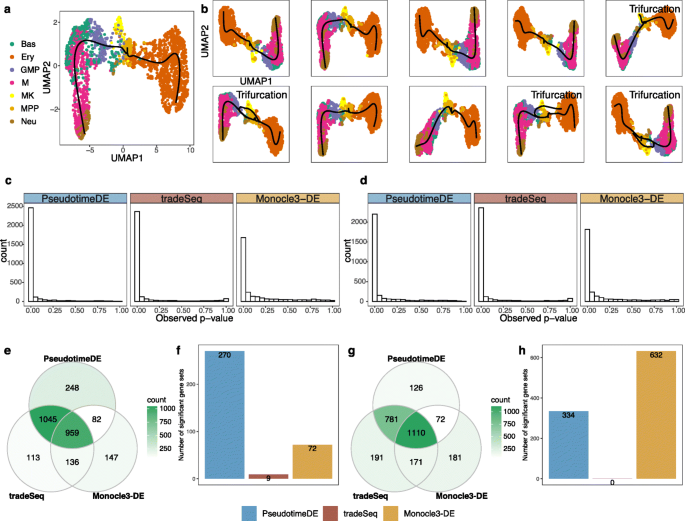
PseudotimeDE: inference of differential gene expression along cell pseudotime with well-calibrated p-values from single-cell RNA sequencing data | Genome Biology | Full Text

PseudotimeDE: inference of differential gene expression along cell pseudotime with well-calibrated p-values from single-cell RNA sequencing data | bioRxiv